
Anders Carlsson has been at Washington University since 1983, and is Professor of Physics. His research interests are in mechanobiology are focused on modeling the polymerization processes underlying the crawling motions of cells. This work is being performed in collaboration with Professor John Cooper at the Washington University School of Medicine. The motions of cells, extensions of their boundaries, and cell division often involve polymerization and depolymerization of the protein actin. This protein forms long, fairly stiff filaments, whose growth can supply the mechanical force necessary for cell motion or shape changes. The filaments are often found in branched or cross-linked networks. The distribution of actin inside cells displays spontaneous dynamic behaviors that are similar to those of an excitable medium. The modeling work treats these phenomena with methods including Brownian-dynamics and stochastic-growth simulations, and analytic theory. The main goals of the modeling are to are to to establish how actin polymerization generates force to "sculpt" the cell membrane in processes such as endocytosis, to pin down the key mechanisms driving migration of cells, to clarify force generation by actomyosin and actin bundles. We are currently expanding our work in mechanobiology to understand how mechanosensitive channels regulate hydration of pollen grains, in collaboration with the Haswell lab at Washington University. This work is supported by the Center for Engineering Mechanobiology.
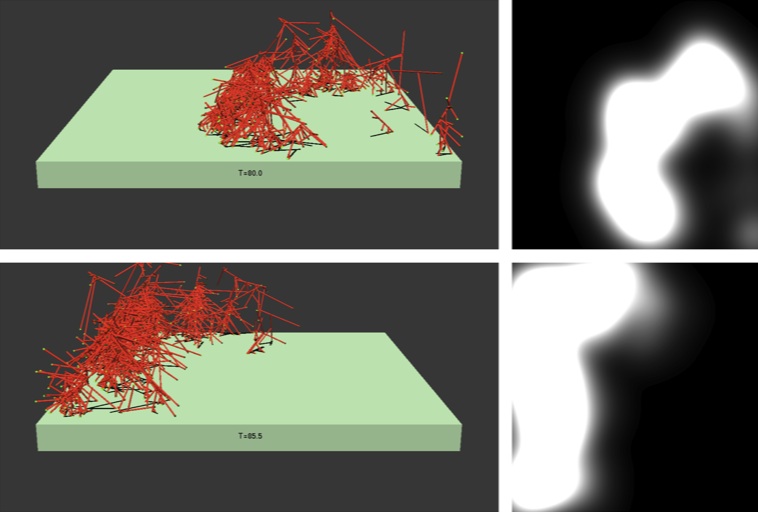
Shown above is the structure of an actin wave propagating across a 3 by 3 micron piece of
cell membrane. Such waves occur in Dictyostelium and several other cell types, and we are
working on pinning down the networks that drive them. Take a look at the movie of simulated
fluorescence .
These results were obtained from a stochastic model of actin filament growth combined
with Brownian dynamics simulations. More recently, under support from NIH
Grant R01 GM107667, we have extended these methods to treat
endocytosis.
